AdK Gromacs Tutorial¶
| Gromacs: | 5.x, 2016, 2018 |
|---|---|
| Tutorial: | 2.0.1 |
| Date: | Jul 03, 2018 |
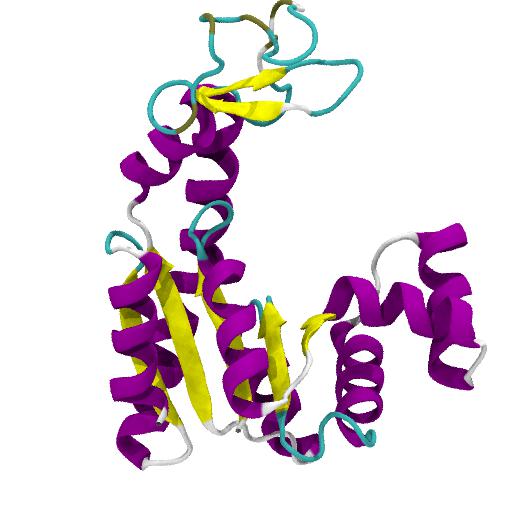
Objective¶
Perform an all-atom molecular dynamics (MD) simulation—using the Gromacs MD package—of the apo enzyme adenylate kinase (AdK) in its open conformation in a physiologically realistic environment, and carry out a basic analysis of its structural properties in equilibrium.
Tutorial files¶
All of the necessary tutorial files can be found on GitHub in the Becksteinlab/AdKGromacsTutorial/tutorial directory, which can be easily obtained by git-cloning the repository:
git clone https://github.com/Becksteinlab/AdKGromacsTutorial.git
Workflow overview¶
For this tutorial we’ll use Gromacs (versions 5, 2016, 2018 should
work) to set up the system, run the simulation, and perform
analysis. An initial structure is provided, which can be found in the
tutorial/templates directory, as well as the MDP files that
are necessary for input to Gromacs. The overall workflow consists of
the following steps: